Metagenomics Unlocks Unknowns of Diarrheal Disease Cases in Children
Nov 19, 2019 — Atlanta, GA
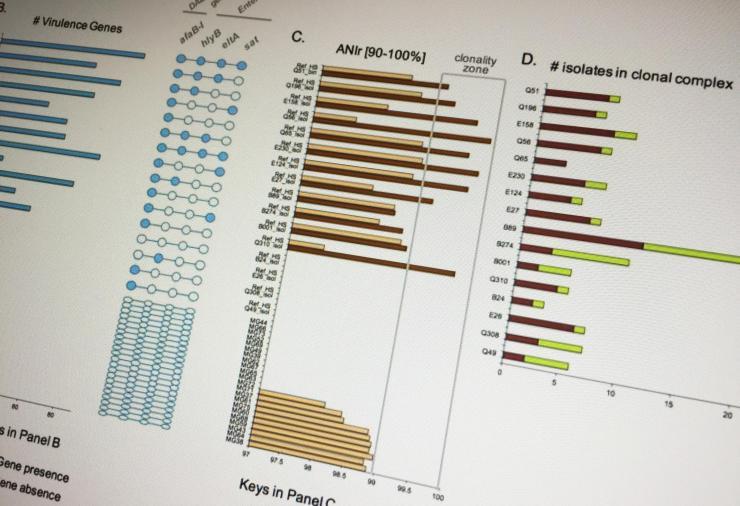
Using advanced metagenomics techniques, researchers have found that conventional culture- based lab tests may often misdiagnose the microbial causes of diarrheal diseases in children. The study, based on samples from Ecuadorian children, also found that a common strain of the E. coli bacterium may be more virulent than previously believed.
Using advanced metagenomics techniques, researchers have found that conventional culture-based lab tests may misdiagnose as many as half of the microbial causes of diarrheal diseases in children. The study, based on samples from Ecuadorian children, also found that a common strain of the E. coli bacterium may be more virulent than previously believed.
The research, which used multiple lines of evidence to determine the microbe causing changes in the gut microbiome during a diarrheal episode, may be significant for applying new diagnostic technologies that could enable personalized medical treatments of intestinal diseases.
The project, led by researchers from the Georgia Institute of Technology, the Emory Rollins School of Public Health, and Universidad San Francisco de Quito, is part of a study funded by the National Institute of Allergy and Infectious Diseases (NIAID) that integrates epidemiological, molecular, and metagenomic data to understand how enteric (food and waterborne) pathogens and the gut microbiome vary across an urban-rural gradient in Ecuador. Frequent illness can affect the growth and development of children during their critical early years.
“We wanted to understand where the illnesses are coming from and what the consequences are for the development of the children,” said Kostas Konstantinidis, a professor in the School of Civil and Environmental Engineering at Georgia Tech. “Knowing more about the causative agent may allow us to prevent infections. This would be relevant not just for the developing world, but for children everywhere.”
The findings were reported Oct. 4 in the journal Applied and Environmental Microbiology. The research team also included researchers from the Universidad Central del Ecuador.
Information the study provided on the likely causative agent of enteric diseases raises questions about long-held definitions of pathogenic agents, said Karen Levy, an associate professor at Rollins. Many so-called “pathogenic E. coli” infections are asymptomatic — not causing diarrhea — she noted. If a microbe considered a pathogen can be detected in a stool sample, but it is not causing disease, is it a pathogen?
“The approach we used in this analysis helps to not only detect whether or not the pathogenic E. coli is present in the stool, but also if it is the likely cause of diarrhea experienced by study subjects,” she said. “This holds promise that, in the future, we could use metagenomic approaches to diagnose not just the presence of the so-called ‘pathogenic E. coli,’ but also of the actual pathogenicity of these organisms.”
Enteric infections often go undiagnosed in children, though they can in some circumstances be fatal. For the study, researchers collected more than 1,000 samples from infected children in Ecuador and analyzed them using traditional culture-based methods in which samples are placed onto selective cell culture media, allowed to grow, and then studied to determine the pathogen present.
A subset of 30 samples were also sent to the Konstantinidis lab at Georgia Tech, where his team has been developing new culture-independent genomics testing that can identify microbes that may not be detectable using standard culture techniques. The researchers examined the total gut microbiome and its shifts during diarrheal infections to assess the effects of three specific pathogenic genotypes of E. coli on the indigenous gut microbiota.
“We looked at the microbes in stool samples using culture-independent genomic techniques,” said Konstantinidis, who also has a faculty position in the Georgia Tech School of Biological Sciences. “We took the DNA out of the samples, sequenced it, and used bioinformatics tools to see what microorganisms were there. By looking at the entire microbiome, we can get more precise information about the pathogens that are causing disease and their effects on the commensal microbes of the gastrointestinal tract: the microbiome. We found that the effects were different for different E. coli pathogens, which is important for diagnosis and distinguishing among them.”
One major finding was that the metagenomics technique disagreed with the culture-based study. “In 50% of the cases where the lab suggested E. coli was the agent, we didn’t see evidence from metagenomics that this was the case,” Konstantinidis said. “We saw evidence that it was something else, potentially an enteric virus.”
The researchers were able to combine different diagnostic approaches in a way that could provide a more complete picture of the microbiome, creating signatures that could potentially identify the microbial agent of the disease. “Compared to the traditional approach, our technique uses multiple lines of evidence, including relative pathogen abundance, population clonality level and detection of virulence factors, and effects on the gut’s natural microbiome that haven’t been used together before,” Konstantinidis said.
The researchers also discovered that a strain known as diffuse adherent E. coli (DAEC) was accompanied by large amounts of co-eluted human DNA in the stool sample. Though DAEC is not generally considered a particularly virulent strain, the presence of human DNA suggests that the DAEC may have been damaging epithelium cells in the gastrointestinal tract.
“Understanding changes in the gut microbiome can help us understand how different strains cause distinct pathogenicity and symptoms such as the elution of high amount of human DNA and changes in the abundance of commensal microbiota,” Konstantinidis said. “If infections like this happen often, that could have implications for a child’s growth and development.”
The metagenomics technique could help provide a foundation for personalized medicine that would select therapies based on the specifics of the microbe – including its virulence and antibiotic resistance profiles. The study shows that the technology works and can be done quickly, but trained personnel are needed to make it more widely available.
“In the next five or 10 years, we are going to see some big changes in which these new methodologies are adopted in the clinic, and that could lead, in the long term, to innovation in diagnosis and treatment,” he added.
Future work as part of another NIAID award will examine how environmental factors – such as water quality, presence of animals in the home, and the chemical environment – affect the microbiomes of small children. In this cohort of children from different geographic areas, the team will examine additional enteric pathogens, and follow children for their first two years of life. This will offer the opportunity to understand whether infections are cleared from the body – or if they continue to lurk in low levels.
In addition to those already mentioned, the authors included recent Georgia Tech School of Biological Sciences graduates Angela Peña-Gonzalez and Maria J. Soto-Girón, Shanon Smith, Jeticia Sistrunk, Lorena Montero, Maritza Páez, Estefanía Ortega, Janet K. Hatt, William Cevallos, and Gabriel Trueba.
Funding for the study was provided by the National Institute of Allergy and Infectious Diseases through grant 1K01AI103544. The contents are solely the responsibility of the author and do not necessarily represent the official views of the NIH.
CITATION: Angela Peña-Gonzalez, et al., “Metagenomic signatures of gut infection caused by different Escherichia coli pathotypes” (Applied and Environmental Microbiology, 2019). https://aem.asm.org/content/early/2019/10/01/AEM.01820-19
Research News
Georgia Institute of Technology
177 North Avenue
Atlanta, Georgia 30332-0181 USA
Media Relations Contact: John Toon (404-894-6986) (jtoon@gatech.edu).
Writer: John Toon
Under high magnification, this digitally colorized scanning electron microscope image depicts a growing cluster of Gram-negative, rod-shaped, Escherichia coli bacteria, of the strain O157:H7, which is a pathogenic strain of E. coli. (Image: Centers for Disease Control and Prevention)
John Toon
Research News
(404) 894-6986




