Periodontitis Bacteria Love Colon and Dirt Microbes
Sep 09, 2019 — Atlanta, GA
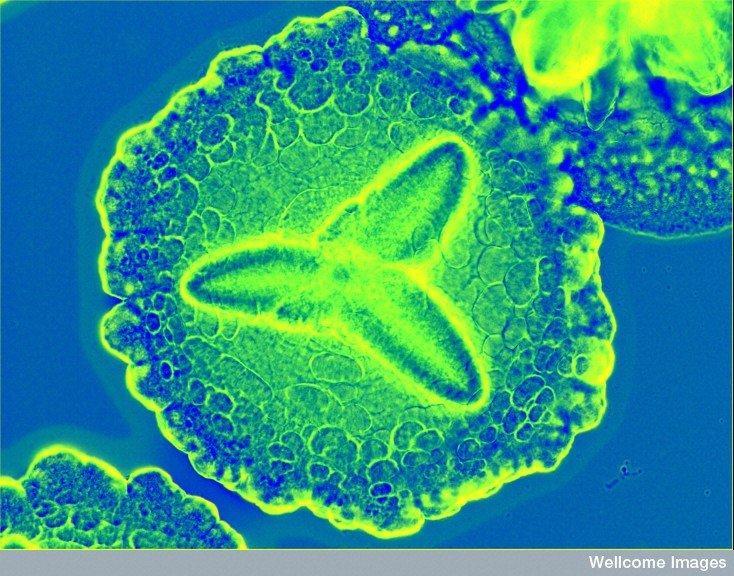
Bacteria behind acute periodontitis. Aggregatibacter actinomycetemcomitans most often lives peacefully in the mouth until circumstances lead it to become infectious. It forms flower-like colonies, here under a microscope sporting a colorful stain added by a researcher. Aa is gram-negative. Credit: Derren Ready (2012) CIL:38942, CIL. Dataset. https://doi.org/doi:10.7295/W9CIL38942 Creative Commons license
True or false? Bacteria living in the same space, like the mouth, have evolved collaborations so generous that they are not possible with outside bacteria. That was long held to be true, but in a new, large-scale study of microbial interactions, the resounding answer was “false.”
Research led by the Georgia Institute of Technology found that common mouth bacteria responsible for acute periodontitis fared better overall when paired with bacteria and other microbes that live anywhere but the mouth, including some commonly found in the colon or in dirt. Bacteria from the oral microbiome, by contrast, generally shared food and assistance more stingily with gum infector Aggregatibacter actinomycetemcomitans, or Aa for short.
Like many bacteria known for infections they can cause – like Strep – Aa often live peacefully in the mouth, and certain circumstances turn them into infectors. The researchers and their sponsors at the National Institutes of Health would like to know more about how Aa interacts with other microbes to gain insights that may eventually help fight acute periodontitis and other ailments.
“Periodontitis is the most prevalent human infection on the planet after cavities,” said Marvin Whiteley, a professor in Georgia Tech’s School of Biological Sciences and the study’s principal investigator. “Those bugs get into your bloodstream every day, and there has been a long, noted correlation between poor oral hygiene and prevalence of heart disease.”
Unnatural pairing
The findings are surprising because bacteria in a microbiome have indeed evolved intricate interactions making it seem logical that those interactions would stand out as uniquely generous. Some mouth microbes even have special docking sites to bind to their partners, and much previous research has tightly focused on their cooperations. The new study went broad.
“We asked a bigger question: How do microbes interact with bugs they co-evolved with as opposed to how they would interact with microbes they had hardly ever seen. We thought they would not interact well with the other bugs, but it was the opposite,” Whiteley said.
The study’s scale was massive. Researchers manipulated and tracked nearly all of Aa’s roughly 2,100 genes using an emergent gene tagging technology while pairing Aa with 25 other microbes — about half from the mouth and half from other body areas or the environment.
They did not examine the mouth microbiome as a whole because multi-microbial synergies would have made interactions incalculable. Instead, the researchers paired Aa with one other bug at a time — Aa plus mouth bacterium X, Aa plus colon bacterium Y, Aa plus dirt fungus Z, and so on.
“We wanted to see specifically which genes Aa needed to survive in each partnership and which ones it could do without because it was getting help from the partner,” said Gina Lewin, a postdoctoral researcher in Whiteley’s lab and the study’s first author. They published their results in the Proceedings of the National Academy of Sciences.
Q & A
How could they tell that Aa was doing well or poorly with another microbe?
The researchers looked at each of Aa’s genes necessary for survival while it infected a mouse -- when Aa was the sole infector, when it partnered with a fellow mouth bacterium and when paired with a microbe from colon, dirt, or skin.
“When Aa was by itself, it needed a certain set of genes to survive – like for breathing oxygen,” Lewin said. “It was striking that when Aa was with this or that microbe that it normally didn’t live around, it no longer needed a lot of its own genes. The other microbe was giving Aa things that it needed, so it didn’t have to make them itself.”
“Interactions between usual neighbors — other mouth bacteria — looked more frugal,” Whiteley said. “Aa needed a lot more of its own genes to survive around them, sometimes more than when it was by itself.”
[Ready for graduate school? Here's how to apply to Georgia Tech.]
How did the emerging genetic marking method work?
To understand “transposon sequencing,” picture a transposon as a DNA brick that cracks a gene, breaking its function. The brick also sticks to the gene and can be detected by DNA sequencing, thus tagging that malfunction.
Every Aa bacterium in a pile of 10,000 had a brick in a random gene. If Aa’s partner bacterium, say, E. coli, picked up the slack for a broken function, Aa survived and multiplied even with the damaged gene, and researchers detected a higher number of bacteria containing the gene.
Aa surviving with more broken genes meant a partner microbe was giving it more assistance. Aa bacteria with broken genes that a partner could not compensate for were more likely to die, reducing their count.
Does this mean the mouth microbiome does not have unique relationships?
It very likely does have them, but the study’s results point to not all relationships being cooperative. Some microbiomes could have high fences and share sparsely.
“One friend or enemy may be driving your behavior, and other microbes may just be standing around,” Lewin said.
Smoking, poor hygiene, or diabetes — all associated with gum disease — might be damaging defensive microbiomes and allowing outside bacteria to help Aa attack gum tissue. It’s too early to know that, but Whiteley’s lab wants to dig deeper, and the research could have implications for other microbiomes.
Also read: Test for Life-Threatening Nutrient Deficit Made From Bacteria Entrails
These researchers coauthored the study: Apollo Stacy from the National Institute of Infectious Diseases and the National Institute of General Medical Sciences, Kelly Michie from Georgia Tech, and Richard Lamont from the University of Louisville. The research was funded by the National Institutes of Health’s National Institute of Infectious Diseases (grants R01DE020100, R01DE023193) and the National Institutes of Health (grants F32DE027281, F31DE024931). Any findings, conclusions or recommendations are those of the authors and not necessarily those of the National Institutes of Health. Whiteley is also a Georgia Research Alliance Eminent Scholar and Co-Director of Emory-Children’s Cystic Fibrosis Center.
Writer & Media Representative: Ben Brumfield (404-660-1408), email: ben.brumfield@comm.gatech.edu
Georgia Institute of Technology
177 North Avenue
Atlanta, Georgia 30332-0181 USA
Graduate research assistant Kelly Michie coauthored the study. Here, she holds up a dish with a bacteria culture. Credit: Georgia Tech / Christopher Moore

Aggregatibacter actinomycetemcomitans breaths oxygen, but in some partnerships with other microbes, it was tested in an anoxic environment. Credit: Georgia Tech / Christopher Moore




