Study Shows How Bacteria Behave Differently in Humans Compared to the Lab
May 22, 2018 — Atlanta, GA
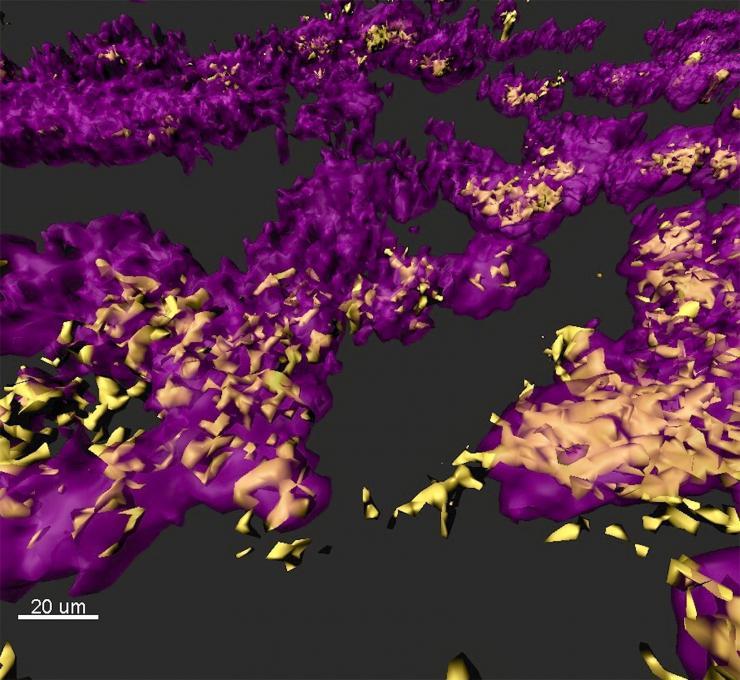
Image shows a rendered confocal laser scanning micrograph of a mixed species bacterial biofilm observed in a chronic wound model of infection. Staphylococcus aureus (yellow) and Pseudomonas aeruginosa (purple), organize themselves in small, dense clusters of cells called aggregates. (Credit: Sophie Darch, Whiteley Lab and Rumbaugh Lab at Texas Tech University Health Sciences Center.)
Most of what we know today about deadly bacteria such as Pseudomonas aeruginosa was obtained from studies done in laboratory settings. Research reported May 14 in the journal Proceedings of the National Academy of Sciences shows that this laboratory-based information may have important limits for predicting how these bugs behave once they’ve invaded humans.
Among the differences are increased expression of genes responsible for antibiotic resistance, the bane of drugs currently used to treat a wide range of infections. The new research could help scientists understand how to draw more accurate conclusions from their laboratory work – and provide doctors with better information on treating bacterial infections.
“Bacteria in human infections are often tolerant of antibiotics, but when we culture them outside the human they are highly susceptible,” said Marvin Whiteley, a professor in the School of Biological Sciences at the Georgia Institute of Technology and co-director of the Emory-Children’s Cystic Fibrosis Center. “In this paper, we show that several genes important for antibiotic tolerance are highly induced in humans compared to our laboratory and mouse modeling systems. There appears to be something unique in the human that is promoting resistance.”
What might be causing that difference remains a mystery, though bacteria are known to be affected by their environment. Understanding how bacterial genes and their expression levels differ in humans could allow researchers to search for laboratory conditions that better mimic the human conditions – and provide better guidance for the use of antibiotics.
“Understanding which antibiotic resistance genes are highly expressed in humans may inform our therapeutic decisions on antibiotic usage,” said Whiteley, who holds the Bennie H. & Nelson D. Abell Chair in Molecular and Cellular Biology at Georgia Tech and is a Georgia Research Alliance Eminent Scholar. “For instance, one might predict antibiotic resistance of an infecting community from gene expression data without the need for culturing microbes in the clinical lab.”
The study was supported by the National Institutes of Health, the Cystic Fibrosis Foundation, and the Lundbeck Foundation. In addition to the Georgia Tech researchers, the research team included scientists at the Texas Tech University Health Sciences Center, the University of Mississippi Medical Center, the University of California, and several clinical and research organizations in Denmark.
Pseudomonas aeruginosa is an important pathogen that threatens immunocompromised people, including those with cystic fibrosis, diabetes and obesity. It is a major hospital-acquired infection, and the Centers for Disease Control and Prevention characterizes multi-drug resistant strains of the bacteria as a serious threat.
In their research, the scientists analyzed RNA sequencing data from both human clinical infections and laboratory experiments. The human samples were obtained from collaborating clinicians, who took the samples directly from patients and put them into a chemical that preserved their RNA for later processing. The laboratory experiments studied different strains of the bacterium under a variety of growth conditions, from antibiotic treatment to competition with other bacteria.
The researchers also included previously published in vitro and mouse experiment data from the Whiteley laboratory and other research teams. Data analysis techniques included a machine learning approach known as Support Vector Machines, which was used to distinguish between gene expression profiles of samples taken from human and in vitro sources.
“We saw high expression in several genes notorious for antibiotic resistance, including genes that encode efflux pumps that extrude antibiotics from the cell as well as an enzyme that degrades certain antibiotics, such as ampicillin,” said Daniel Cornforth, a research scientist in Whiteley’s laboratory and the paper’s first author. “There were also less studied antibiotic resistance genes, including three related to zinc transport that our previous work has identified as critical antibiotic resistance determinants that were also highly expressed in the human patients.”
Though the research focused only on a single troublesome pathogen, Whiteley believes the results could have broader implications. “We actually know very little about bacteria behaviors during human infection and most model systems cannot replicate most aspects of human infection. I expect that this work would be generalizable to other bacteria.”
By identifying how bacteria behave differently in humans compared to standard laboratory settings, the work could provide a foundation for additional study with more samples and different types of infection.
“The key takeaway from this work is that now microbiologists can perform transcriptomics on bacterial populations in a range of human infections, so we can better understand what bacteria are actually doing in these clinical infections,” said Cornforth. “We can also determine where our laboratory models succeed and where they fail in mimicking these infection environments.”
This study was funded by National Institutes of Health Grant R01GM116547-01A1, a Human Frontiers Science grant, Cystic Fibrosis Foundation Grant WHITEL16G0, Lundbeck Foundation Grant R204-2015-4205 and Lundbeck Foundation Grant R105-A9791, and by Cystic Fibrosis postdoctoral Fellowships CORNFO15F0 and IBBERS16F0.
CITATION: Daniel Cornforth, et al., “Pseudomonas aeruginosa transcriptome during human infection,” (Proceedings of the National Academy of Sciences, 2018). https://doi.org/10.1073/pnas.1717525115
Research News
Georgia Institute of Technology
177 North Avenue
Atlanta, Georgia 30332-0181 USA
Media Relations Contact: John Toon (404-894-6986) (jtoon@gatech.edu).
Writer: John Toon

Research Scientist Dan Cornforth and Postdoctoral Fellow Carolyn Ibberson are shown in the laboratory of Professor Marvin Whiteley at Georgia Tech. They are part of a research team studying differences in how bacteria behave in laboratory conditions compared to in human infections. (Credit: Rob Felt, Georgia Tech)

Research Scientist Dan Cornforth and Postdoctoral Fellow Carolyn Ibberson are shown in the laboratory of Professor Marvin Whiteley at Georgia Tech. They are part of a research team studying differences in how bacteria behave in laboratory conditions compared to in human infections. (Credit: Rob Felt, Georgia Tech)
John Toon
Research News
(404) 894-6986




