Whole-brain Functional Imaging Takes New Leaps with Deep Learning
Oct 03, 2022 — Atlanta, GA
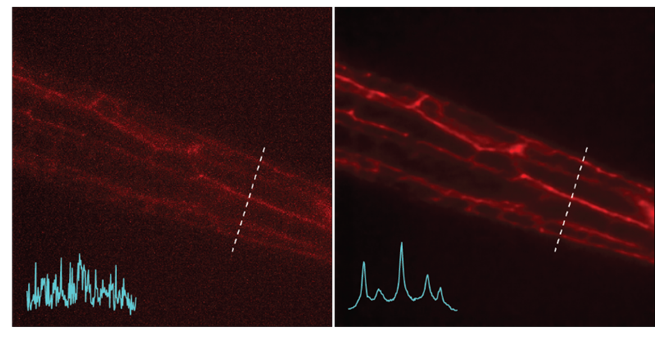
Left: Noisy images showing neuronal structures. Right: NIDDL Deep Denoised image of neuronal structures.
Imaging neuro activities for long durations, high speeds, and large regions of the brain simultaneously is critical to understand the underlying computations in the brain – in normal functions as well as in developmental and neurodegenerative diseases, according to Georgia Tech researchers.
Model organisms in neuroscience such as the round worm C. elegans enable these kinds of studies to be conducted. This is because researchers can record activities of their entire brains simultaneously with new microscopy techniques. However, collecting such data is difficult with commonly available setups due to several technical constraints.
But a new study published in Nature Communications shows that deep learning (a machine-learning technique) can overcome technical constraints in whole-brain imaging, enabling new experiments that were previously not possible. Furthermore, this method will enable every lab in the world with common microscopy setups to do whole-brain imaging and accelerate discovery.
The study’s authors – Professor Hang Lu, Cecil J. “Pete” Silas Chair in Georgia Tech’s School of Chemical and Biomolecular Engineering (ChBE) and Shivesh Chaudhary, ChBE PhD 2022 – are interested in uncovering the fundamental building blocks of intelligence. For this, they focus on the nervous system of C. elegans, small and compact with only 302 cells, but capable of generating complex behaviors.
Technical Constraints
Whole brain optical imaging methods allows researchers to record neuron activities at single-cell resolution, providing unprecedented amounts of data. And yet, commonly available confocal microscopes are not able to handle the technical constraints required for whole brain imaging, the researchers said.
For example, the imaging must be performed at fast speeds to capture neuron activity dynamics, and to minimize motion artifacts. In addition, laser power must be minimized to prevent photobleaching of fluorescent markers used to label neurons, and to avoid photo-toxicity to animals.
Because of these constraints, the videos can be extremely noisy. As a result, several critical downstream processing tasks, such as cell detection and tracking, become extremely complicated, and neuron activity extracted from these videos is of poor quality.
Deep Learning Strategies
To overcome these challenges, the research team wondered if they could use advances in deep learning methods to reduce noise from lower quality but experimentally accessible imaging techniques.
In principle, this can be done in two ways. First, using a very large collection of data, unsupervised methods (not requiring human monitoring) can be “taught” to learn about features in the noisy images that can be turned into signals of interest to researchers.
Supervised deep learning, in comparison, would take advantage of what the signal-enhanced images should ideally look like and how the corresponding noisy images appear, instructing the machine what it’s supposed to see.
“Supervised deep learning uses a smaller set of data to teach the machine what to look for, and with the right set of data, is really good at the task,” Lu explained.
But both unsupervised and supervised deep learning approaches pose some significant challenges for the whole-brain imaging problem – the number of training datasets for unsupervised methods is insurmountably large, and the training datasets for traditional supervised methods are technically infeasible in that it is not possible to obtain the high-signal (high resolution) and noisy images that have the exact correspondence.
Developing New Framework
To solve these technical challenges, the research team designed a new supervised deep learning method called Neuro-Imaging Denoising via Deep Learning (NIDDL).
The NIDDL framework demonstrates advantages compared to previous works. The method simplifies training data collection strategy because the network can be trained with still images consistent with neuro activity captured on video.
To explain, Professor Lu uses an analogy of processing a blurry video of someone playing tennis. Traditional algorithms would require one to use exact shots for comparison (one blurry, one not), but that’s not doable because the camera cannot provide both images at once. The tennis player would have moved by the time of the second photo, no matter how fast the exposure. But if the tennis player posed a position, then you could take two images and use them to train the algorithm.
With C. elegans in place of the tennis player, the researchers can use still shots of sedated organisms for training the deep learning algorithm, Lu said.
Thus, unlike in previous studies, ultrafast imaging rates for training data collection are not required. In addition, NIDDL framework requires much lower training data (approximately 500 pairs of images) compared to previous methods that require 3000-30,000 frames, because this deep learning technique has streamlined the process.
NIDDL can be trained in supervised fashion across images of varieties of strains, labelling markers, and noise levels, making them more generalizable.
Adoptable Technology
The manageable amount data required for training could encourage labs to set up their own denoising pipelines using the framework, the researchers said.
Also, more labs could adopt the NIDDL framework without having access to powerful graphic processing units. NIDDL has been extensively optimized to achieve a 20 to 30 times smaller memory footprint and a three to four times faster inference time (the amount needed for the machine to make a prediction) compared to previous methods.
“Labs could run experiments faster, easier, cheaper, and better,” Lu said.
Future Applications
The researchers believe that NIDDL can be applied to many neural imaging scenarios, where experimentalists would only need to curate a small set of data specific to their experiments to deploy the algorithm to denoise the data.
For example, NIDDL would enable researchers to circumvent some experimental bottlenecks to make faster recordings to resolve brain dynamics, longer recordings to study a variety of behaviors in animals, and cover larger brain areas.
“These applications would push forward our fundamental understandings of how the brain works, and guide understandings of brain disease mechanisms and discovering therapeutics,” Chaudhary said.
CITATION: https://www.nature.com/articles/s41467-022-32886-w
FUNDInG: The authors acknowledge the funding support of the U.S. NIH (R01NS096581, R01MH130064, and R01NS115484) and the U.S. NSF (1764406 and 1707401) to H.L. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the NIH (P40 OD010440), National Center for Research Resources and the International C. elegans Knockout Consortium. This research was supported in part through research cyberinfrastructure resources and services provided by the Partnership for an Advanced Computing Environment (PACE) at the Georgia Institute of Technology, Atlanta, Georgia, USA.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the view of the sponsoring agency.
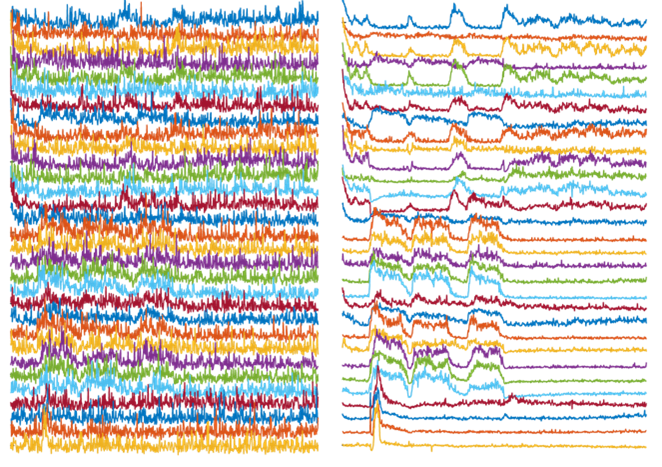
Left: Activity traces of neurons from noisy videos, with noise masking real activities of interest. Right: Activity traces of neurons from NIDDL Deep Denoised videos unmasking the activities of interest.
Brad Dixon, braddixon@gatech.edu




